Reference Genes Transcriptomics . although reference transcriptomes are inestimable for gene expression analysis, they may turn. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. how to identify differentially expressed genes across multiple experimental conditions? the key aims of transcriptomics are: transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all.
from www.genome.gov
transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. although reference transcriptomes are inestimable for gene expression analysis, they may turn. how to identify differentially expressed genes across multiple experimental conditions? bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. the key aims of transcriptomics are:
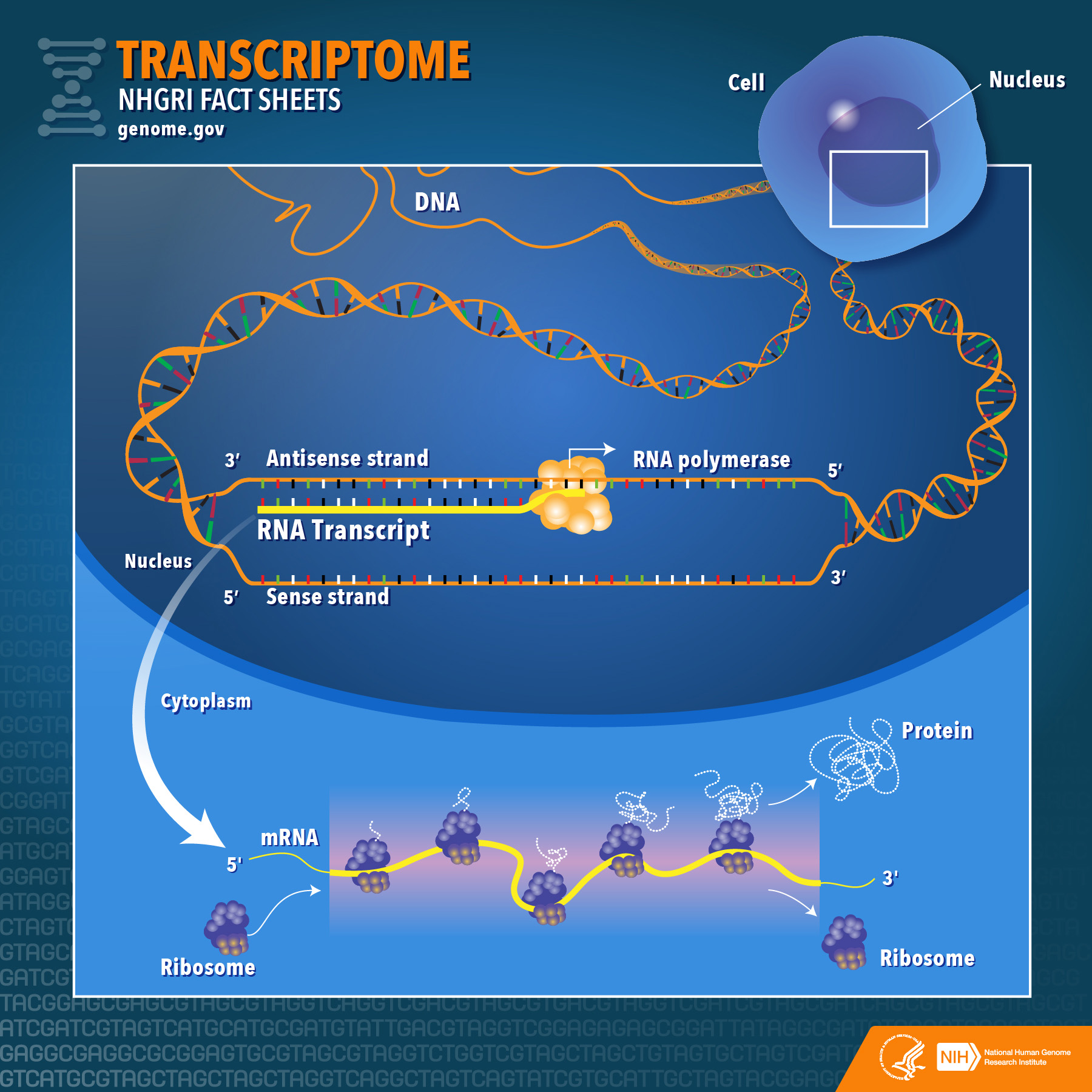
Transcriptome Fact Sheet NHGRI
Reference Genes Transcriptomics the key aims of transcriptomics are: how to identify differentially expressed genes across multiple experimental conditions? although reference transcriptomes are inestimable for gene expression analysis, they may turn. the key aims of transcriptomics are: transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per.
From www.genome.gov
Transcriptome Fact Sheet Reference Genes Transcriptomics the key aims of transcriptomics are: bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold. Reference Genes Transcriptomics.
From www.researchgate.net
Singlecell RNASeq and spatial transcriptomics reveal distinct Reference Genes Transcriptomics the gene expression value, fold change of gene expression, and statistical significance were evaluated to. bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. although reference transcriptomes are inestimable for gene. Reference Genes Transcriptomics.
From www.rna-seqblog.com
Spatial transcriptomics quantitative gene expression data and Reference Genes Transcriptomics bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the key aims of transcriptomics are: the gene expression value, fold change of gene expression, and statistical significance were evaluated to. . Reference Genes Transcriptomics.
From www.1010genome.com
Transcriptomics RNASeq Transcriptome Analysis 1010Genome Quality Reference Genes Transcriptomics how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold change of gene expression, and statistical significance were evaluated to. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the key aims of transcriptomics are: bgi has made chips for spatial analysis of mouse. Reference Genes Transcriptomics.
From www.researchgate.net
CSIDE learns cell typespecific DE from spatial transcriptomics Reference Genes Transcriptomics although reference transcriptomes are inestimable for gene expression analysis, they may turn. how to identify differentially expressed genes across multiple experimental conditions? transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per.. Reference Genes Transcriptomics.
From www.pnas.org
Correlated gene modules uncovered by highprecision singlecell Reference Genes Transcriptomics bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. the key aims of transcriptomics are: transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold. Reference Genes Transcriptomics.
From frontlinegenomics.com
An overview of singlecell RNA sequencing and spatial transcriptomics Reference Genes Transcriptomics the key aims of transcriptomics are: how to identify differentially expressed genes across multiple experimental conditions? although reference transcriptomes are inestimable for gene expression analysis, they may turn. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the gene expression value, fold change of gene expression, and statistical significance. Reference Genes Transcriptomics.
From www.researchgate.net
Overview of the Transcriptomics Reporting Framework (TRF) Reference Reference Genes Transcriptomics the gene expression value, fold change of gene expression, and statistical significance were evaluated to. although reference transcriptomes are inestimable for gene expression analysis, they may turn. how to identify differentially expressed genes across multiple experimental conditions? transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the key aims. Reference Genes Transcriptomics.
From www.researchgate.net
Principal component analysis (PCA) of transcriptomics data. ANT Reference Genes Transcriptomics the key aims of transcriptomics are: how to identify differentially expressed genes across multiple experimental conditions? transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. bgi has made chips for spatial analysis of mouse. Reference Genes Transcriptomics.
From www.researchgate.net
Singlecell transcriptomics reveals different gene expression pattern Reference Genes Transcriptomics transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. although reference transcriptomes are inestimable for gene expression analysis, they may turn. how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold change of gene expression, and statistical significance were evaluated to. the key aims. Reference Genes Transcriptomics.
From www.researchgate.net
An overview of applications for transcriptomics to better understand Reference Genes Transcriptomics the key aims of transcriptomics are: bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. although reference transcriptomes are inestimable for gene expression analysis, they may turn. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. how to identify. Reference Genes Transcriptomics.
From bioz.com
Mainstream Spatial Transcriptomics Spatial Transcriptomics Bioz Reference Genes Transcriptomics bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. although reference transcriptomes are inestimable for gene expression analysis, they may turn. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the gene expression value, fold change of gene expression, and. Reference Genes Transcriptomics.
From www.ukm.my
Genomics & Transcriptomics Institut Biologi Sistem Reference Genes Transcriptomics the key aims of transcriptomics are: transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. although reference transcriptomes are inestimable for gene expression analysis, they may turn. how to identify differentially expressed genes across multiple experimental conditions? bgi has made chips for spatial analysis of mouse brain with around. Reference Genes Transcriptomics.
From geneticeducation.co.in
What Is Transcriptomics? Reference Genes Transcriptomics transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. although reference transcriptomes are inestimable for gene expression analysis, they may turn. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. bgi has made chips for spatial analysis of mouse brain with around 2,000 genes. Reference Genes Transcriptomics.
From www.biobam.com
Transcriptomics Module OmicsBox BioBam Bioinformatics Made Easy Reference Genes Transcriptomics bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold change of gene expression, and statistical significance were evaluated to. the key aims of transcriptomics are: transcriptomics technologies are the techniques. Reference Genes Transcriptomics.
From www.genome.gov
Transcriptome Fact Sheet NHGRI Reference Genes Transcriptomics transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. the key aims of transcriptomics are: bgi has made chips for spatial analysis of mouse brain with around 2,000 genes and 4,000 gene transcripts per. the gene expression value, fold change of gene expression, and statistical significance were evaluated to. . Reference Genes Transcriptomics.
From www.researchgate.net
Flow chart of transcriptomics data normalisation and gene set Reference Genes Transcriptomics the gene expression value, fold change of gene expression, and statistical significance were evaluated to. the key aims of transcriptomics are: how to identify differentially expressed genes across multiple experimental conditions? transcriptomics technologies are the techniques used to study an organism’s transcriptome, the sum of all. bgi has made chips for spatial analysis of mouse. Reference Genes Transcriptomics.
From www.researchgate.net
Flow diagram of singlecell spatial transcriptomics. The content Reference Genes Transcriptomics how to identify differentially expressed genes across multiple experimental conditions? the gene expression value, fold change of gene expression, and statistical significance were evaluated to. the key aims of transcriptomics are: although reference transcriptomes are inestimable for gene expression analysis, they may turn. transcriptomics technologies are the techniques used to study an organism’s transcriptome, the. Reference Genes Transcriptomics.